Catalog of Regulatory Science Tools to Help Assess New Medical Devices
sFRC (scanning Fourier Ring Correlation) is a tool that compares radiological images from AI or iterative-based image restoration algorithms against those from standard-of-care analytical algorithms to identify and label hallucinations (aka fakes) using small red bounding boxes, which serve as visual indicators of the detected hallucinations.
Technical Description
AI-based methods are currently being explored to restore images from sparse-view, low-resolution, and under-sampled acquisitions in radiology. Although AI-restored images may appear visually appealing based on subjective criteria (such as less noise and smooth features), they may also suffer from readily non-discernable hallucinations (aka fakes). Hallucinations (either additive with respect to the addition of false structures or subtractive with respect to the removal of true structures) in AI-restored images from the subsampled acquisition are very difficult for human eyes to discern, mainly because hallucinations disguise themselves as real anatomy even when they are not present in a patient's internal organs. Easy-to-use techniques and robust metrics are needed to identify hallucinations in AI-based outputs.
The sFRC tool features an image-processing-based solution to automatically and objectively evaluate hallucinations in AI-restored images. This method involves comparing small patches between reference and AI-restored images using the Fourier ring correlation (FRC) analysis method in a (s)canning fashion. Hence, the methodology is termed as sFRC. The sFRC's threshold for hallucination detection is set based on a few pre-identified small regions of interest (ROIs) with known hallucinations. We have demonstrated the effectiveness of sFRC in finding hallucinations in various scenarios, including CT super-resolution and MRI-based under-sampled acquisition. Additionally, we have shown its ability to capture decay in AI performance in out-of-distribution data compared to in-distribution data. sFRC has also been demonstrated to identify hallucinations when iterative (IRT) regularization-based methods are used to reconstruct images from subsampled MRI acquisitions. A summary of the method, input, and output are provided below.
- Method: This software allows users to scan and perform sFRC analysis using pairs of small-sized patches from AI or iterative-based image restoration methods and their reference counterparts. A user-defined hallucination threshold – based on clinical criteria of acceptable image quality or based on imaging theory – is repeatedly applied when scanning across patches in each test image to detect locally limited hallucinations. The reference images are sequestered using analytical standard-of-care methodology. For example, in CT, the reference images can be filtered back projection (FBP) reconstructed images acquired at a normal-dose and at a standard resolution. In MRI, the reference images can be inverse Fourier Transform (iFT) reconstructed images from fully sampled k-space data.
- Input: Restored medical images using AI or iterative-based methods and their reference counterparts from the standard-of-care techniques and a hallucination threshold. The hallucination threshold is used to judge whether or not an sFRC curve corresponding to a patch is hallucinated. The hallucination threshold can be directly input by users or set using a few patches or ROIs that are predefined as hallucination by human observers or is determined using imaging theory-based limitation for a given undersampled image restoration problem [1, 2].
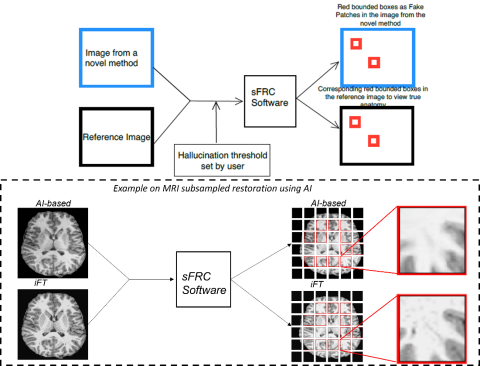
Fig 1: An illustration of inputs to sFRC as test images from novel methods (such as AI or Iterative-based) and reference methods (such as analytical-based), and hallucination threshold. The figure also depicts outputs as red-bounding boxes on images from the novel and reference methods to indicate hallucinated patches and actual anatomy in the corresponding reference patches. The bottom plot depicts an example of using sFRC for a subsampled MRI restoration problem. The novel method is AI-based, and the reference method is inverse Fourier transform (iFT). From the zoomed patches we can explicitly see the removal of a dark signal in the AI-based image as compared to the iFT image.
- Output: Small-sized red bounding boxes on input images deemed as hallucinated ROIs in AI-assisted (or IRT-based) images and in reference images, and the total number of such hallucinated ROIs in the supplied input images. The folder path where the AI-based or IRT-based images with hallucinated ROIs are saved is also displayed as command line output.
As red bounded boxes, these ROIs indicate that those regions have not been faithfully reconstructed. They may exhibit imaging errors that are readily non-discernible hallucinations to human eyes. These fakes or hallucinations may include over-smoothing, in-homogeneity, tiny structural changes, removal of subtle features or signals, distortion of small organelles, the addition of minute indentations, blood vessels, or plaque-like structures, coalescence of tiny organelles, unwarranted folding, contrast migration anomalies, etc.
- Github repository: The codes to perform sFRC analysis as a software package have been compiled into a GitHub repository (https://github.com/DIDSR/sfrc). This repository includes a user manual on how to install software. It also includes example codes on how to execute the software using MR and CT images from AI-based and iterative-based algorithms to find hallucinations.
Intended Purpose
The sFRC software is intended to objectively and automatically identify hallucinations or fakes when AI-based or iterative-based restoration algorithms (either image reconstruction from raw data or image postprocessing) are used to produce images—for modalities like CT and MRI—from subsampled data acquisition processes (such as sparse-view, low-resolution, accelerated acquisition). This software is applicable, but not limited to the assessment of performance of devices during the product development phase which may be submitted for a 510k clearance under the following product codes: LNH, JAK, LLZ.
sFRC is intended to help establish whether the image quality of restored outputs from AI—or iterative-based algorithms for time or dose-saving applications through a subsample data acquisition procedure is hallucinatory compared to the standard-of-care analytical methods under fully sampled acquisition.
Intended users are:
- CT and MRI device developers.
- CT and MRI image reconstruction developers.
- CT and MRI postprocessing software developers.
Testing
Code validation was performed to ensure that sFRC functions as designed. Specifically, for the patches labeled as hallucinations and the total number of hallucinatory patches when sFRC is applied on AI- or iterative-based methods:
- Patches labeled as hallucinations by sFRC were demonstrated to overlap fully with the regions labeled as hallucinations obtained using a separate imaging theory-based methodology [1, 2]. This validation was performed for the MRI subsampling problem using AI and iterative-based methods on the T1w pediatric epilepsy resection dataset [3].
- sFRC-based outcome on the number of hallucinations was demonstrated to subscribe to the commonly known fact that AI algorithms underperform on out-of-distribution test sets compared to in-distribution test sets [4, 5]. This was demonstrated for a CT super-resolution problem using adult CT images provided in the Low-dose Grand challenge [2, 6].
- sFRC-based outcome on the number of hallucinations was demonstrated to subscribe to the data processing inequality, which states that no more information can be obtained out of a set of data than was there to begin with [7]. This was demonstrated for MRI subsampling problem using AI- as well as iterative-based methods on T1w pediatric epilepsy resection dataset. Importantly, sFRC-based hallucinations were shown to increase sequentially in accordance with the increase in subsampling rate [2].
- sFRC was executed using CT and MRI data for a wide range of processers (e.g., 1, 2, 16, 36) to distribute the computation for the same testing data set. As expected, the total number of hallucinations and patches labeled as hallucinations remained the same when results using different number of processors were compared.
- In coordination with two medical officers, we verified that the patches labeled as hallucinations obtained using sFRC truly exhibited imaging errors when AI- or iterative-based methods were used to restore images from subsampled or low-resolution data acquisition processes. These errors include over-smoothing, in-homogeneity, tiny structural changes, removal of subtle features/signals, distortion of small organelles, the addition of minute indentation-/blood vessel-/plaque-like structures, coalescing of tiny organelles, unwarranted folding, contrast migration anomaly, etc. [2].
Limitations
- Similar to other fidelity metrics like mean squared error (MSE) or structure similarity index measure (SSIM), sFRC needs reference images that are counterparts of AI-based images obtained using an analytical reconstruction algorithm that is the current standard-of-care method such as Filtered Back Projection method under a full-dose scan in CT. It cannot determine hallucinations on the fly without reference images.
- sFRC is not a tool to detect deep fakes from generative models for computer vision-related application. Neither is it a tool to detect text-based hallucination from Large Language models.
- sFRC has the potential to find hallucinations when AI-based domain transfer models are used in whole slide imaging-based applications such as color harmonization or color transfer and radiotherapy-based applications such as transferring images from MR to CT. However, sFRC has only been tested and validated for restoration applications like super-resolution and subsampling. It has yet to be validated for domain transfer-based applications.
- A clinically relevant hallucination threshold has to be set using clinically predefined criteria on what is deemed to be hallucinations ROIs by human observers (such as medical officers, imaging scientists), or imaging theory. Note that the clinically relevant hallucination threshold can be different for different anatomical images and imaging modalities.
Supporting Documentation
- Scientific details about the software can be found in the following FDA-cleared preprint. 10.36227/techrxiv.171259560.02243347/v2.
- User Manual with installation, usage instructions, and demo execution codes/guides.
References:
- Bhadra, S., Kelkar, V. A., Brooks, F. J., & Anastasio, M. A. (2021). On hallucinations in tomographic image reconstruction. IEEE transactions on medical imaging, 40(11), 3249-3260.
- Kc, P., Zeng, R., Soni, N., & Badano, A. (2024). sFRC for assessing hallucinations in medical image restoration. Authorea Preprints.
- Maallo, A. M. S., Freud, E., Liu, T. T., Patterson, C., & Behrmann, M. (2020). Effects of unilateral cortical resection of the visual cortex on bilateral human white matter. NeuroImage, 207, 116345.
- Voter, A. F., Larson, M. E., Garrett, J. W., & Yu, J. P. (2021). Diagnostic accuracy and failure mode analysis of a deep learning algorithm for the detection of cervical spine fractures. American Journal of Neuroradiology, 42(8), 1550-1556.
- Zech, J. R., Badgeley, M. A., Liu, M., Costa, A. B., Titano, J. J., & Oermann, E. K. (2018). Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLoS medicine, 15(11), e1002683.
- McCollough, C. H., Bartley, A. C., Carter, R. E., Chen, B., Drees, T. A., Edwards, P., ... & Fletcher, J. G. (2017). Low‐dose CT for the detection and classification of metastatic liver lesions: results of the 2016 low dose CT grand challenge. Medical physics, 44(10), e339-e352.
- McDonnell, M. D., Stocks, N. G., Pearce, C. E., & Abbott, D. (2003, May). The data processing inequality and stochastic resonance. In Noise in Complex Systems and Stochastic Dynamics (Vol. 5114, pp. 249-260). SPIE.
Contact
Tool Reference
- RST Reference Number: RST24MD16.01
- Date of Publication: 09/18/2025
- Recommended Citation: U.S. Food and Drug Administration. (2025). sFRC for Detecting Hallucinations in Medical Image Restoration (RST24MD16.01). https://cdrh-rst.fda.gov/sfrc-detecting-hallucinations-medical-image-restoration
